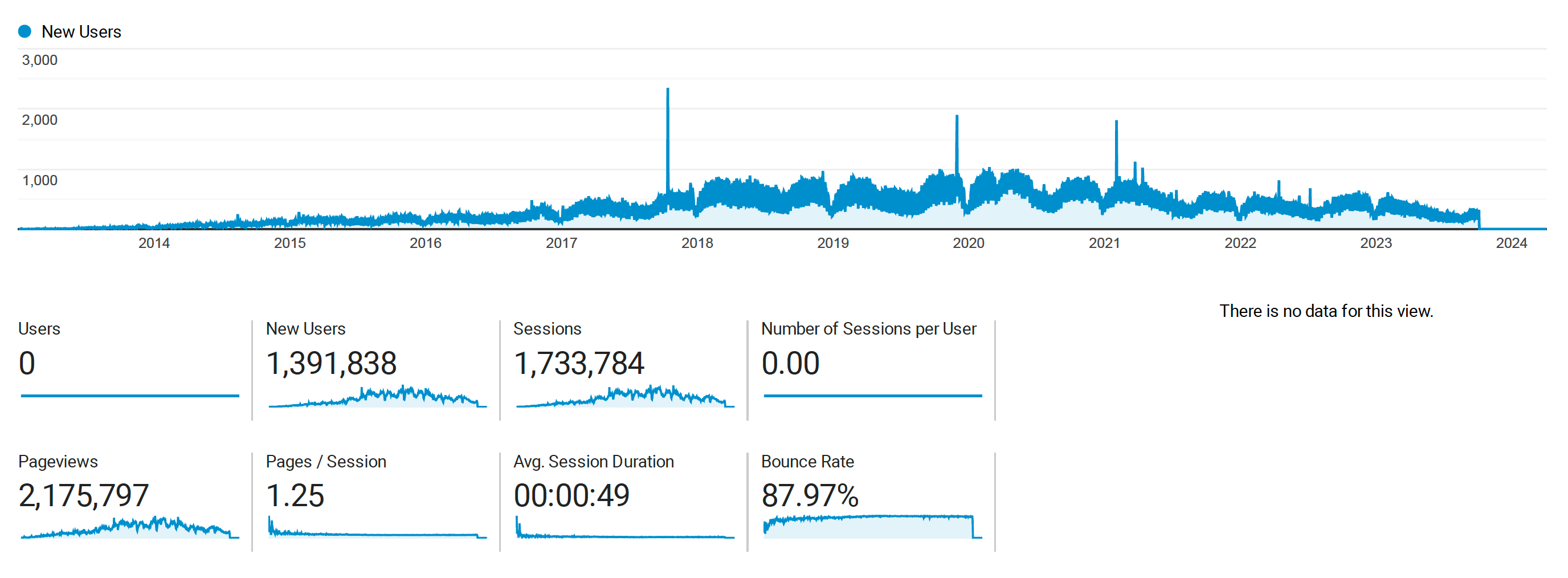
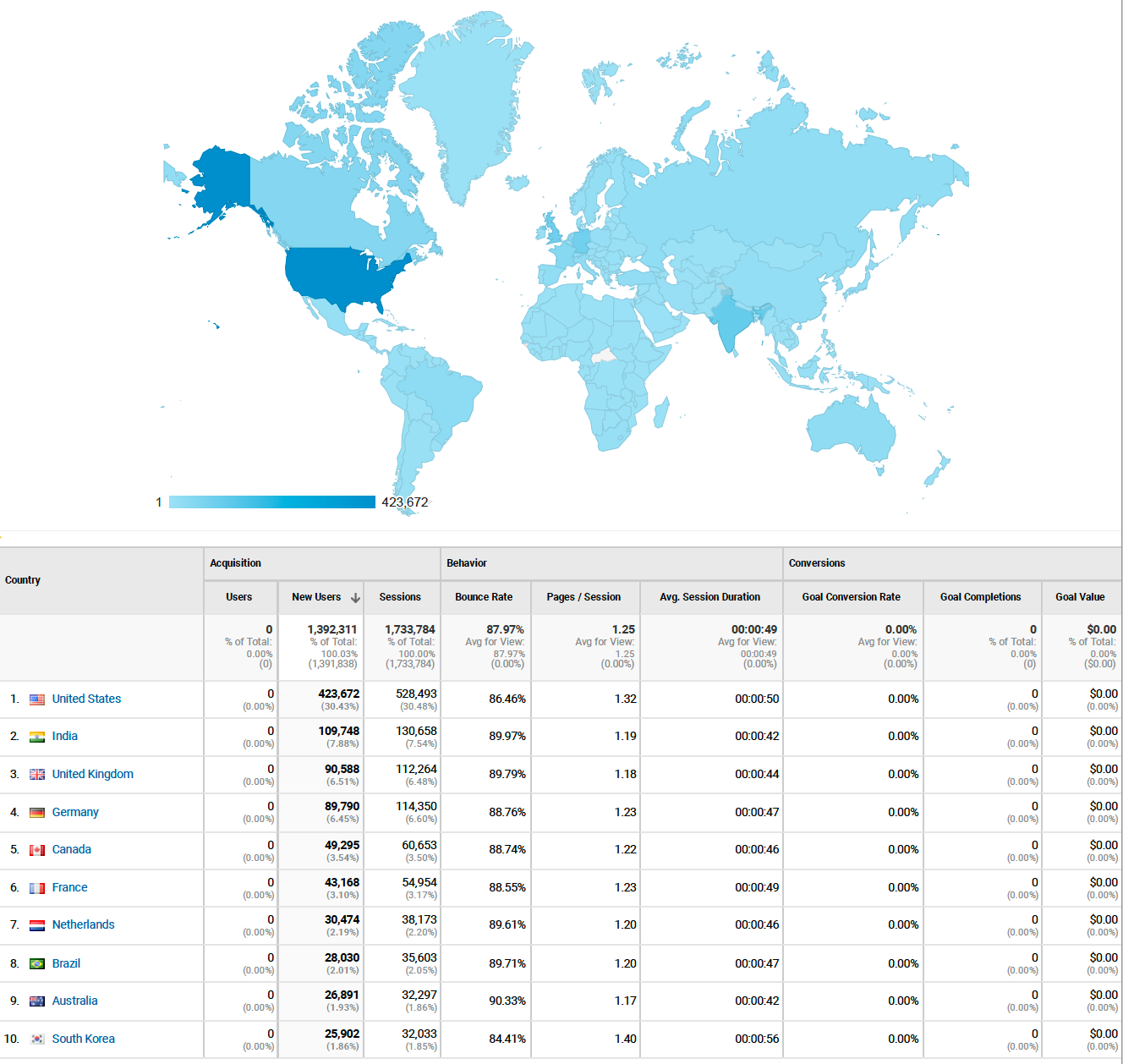
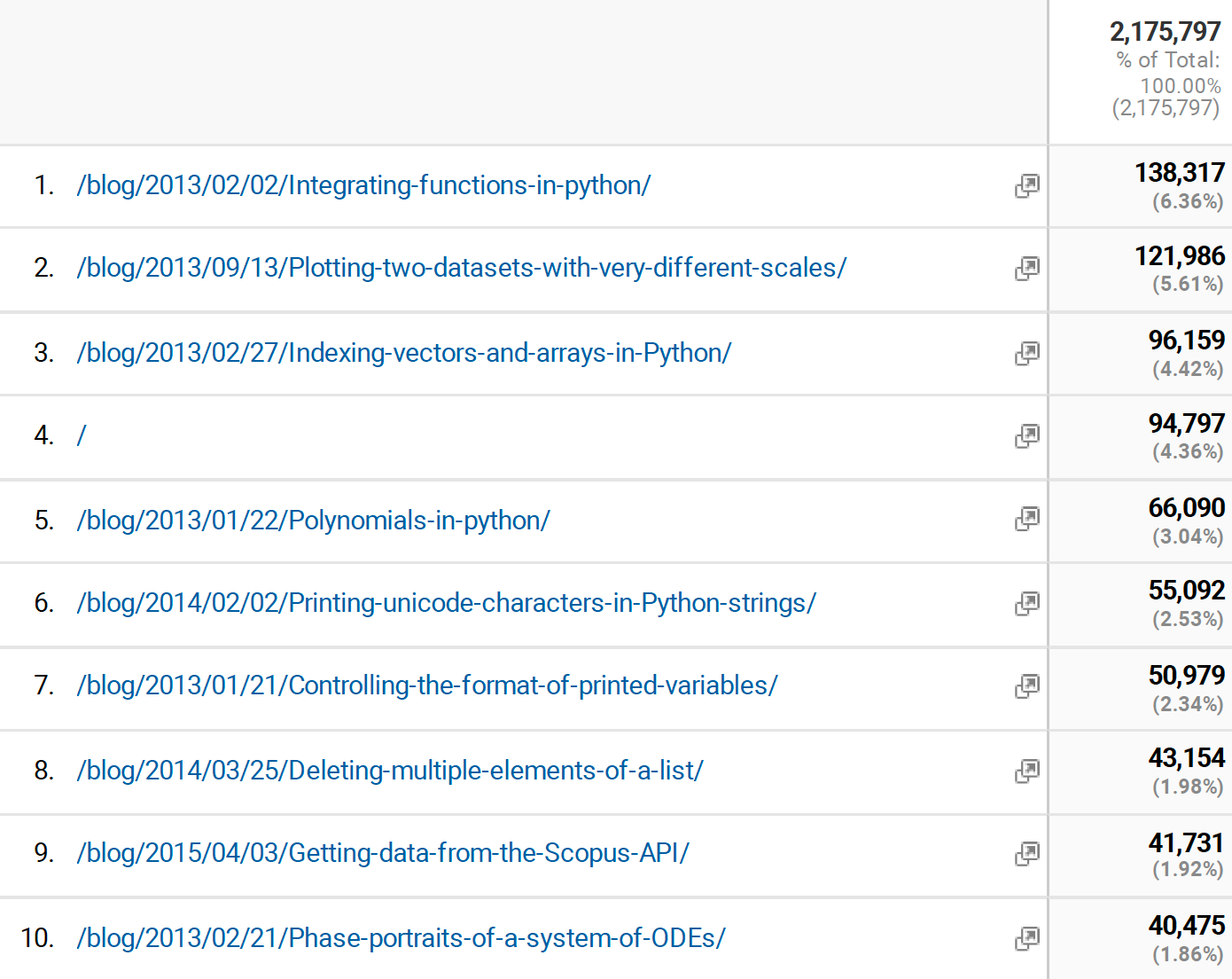
Kolmogorov-Arnold Networks (KANs) and Lennard Jones
Posted May 05, 2024 at 11:06 AM | categories: uncategorized | tags:
Table of Contents
KANs have been a hot topic of discussion recently (https://arxiv.org/abs/2404.19756). Here I explore using them as an alternative to a neural network for a simple atomistic potential using Lennard Jones data. I adapted this code from https://github.com/KindXiaoming/pykan/blob/master/hellokan.ipynb.
TL;DR It was easy to make the model, and it fit this simple data very well. It does not extrapolate in this example, and it is not obvious what the extrapolation behavior should be.
1. Create a dataset
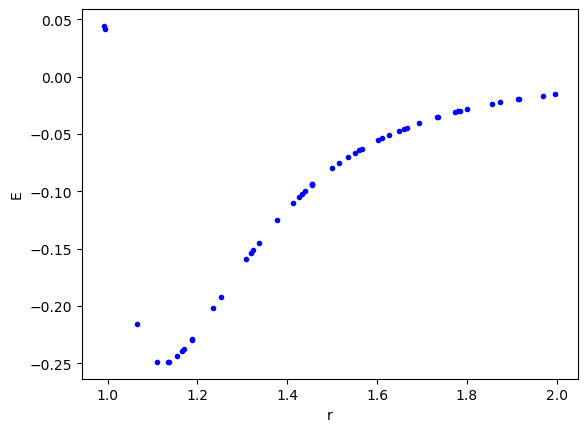
We leverage the create_dataset function to generate the dataset here. I chose a range with some modest nonlinearity, and the minimum.
import matplotlib.pyplot as plt import torch from kan import create_dataset, KAN def LJ(r): r6 = r**6 return 1 / r6**2 - 1 / r6 dataset = create_dataset(LJ, n_var=1, ranges=[0.95, 2.0], train_num=50) plt.plot(dataset['train_input'], dataset['train_label'], 'b.') plt.xlabel('r') plt.ylabel('E');
2. Create and train the model
We start by making the model. We are going to model a Lennard-Jones potential with one input, the distance between two atoms, and one output. We start with a width of 2 "neurons".
model = KAN(width=[1, 2, 1])
Training is easy. You can even run this cell several times.
model.train(dataset, opt="LBFGS", steps=20);
model.plot()
train loss: 1.64e-04 | test loss: 1.46e-02 | reg: 6.72e+00 : 100%|██| 20/20 [00:03<00:00, 5.61it/s]
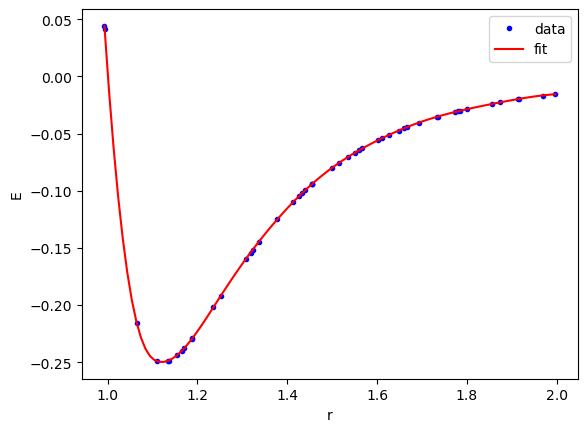
We can see here that the fit looks very good.
X = torch.linspace(dataset['train_input'].min(), dataset['train_input'].max(), 100)[:, None] plt.plot(dataset['train_input'], dataset['train_label'], 'b.', label='data') plt.plot(X, model(X).detach().numpy(), 'r-', label='fit') plt.legend() plt.xlabel('r') plt.ylabel('E');
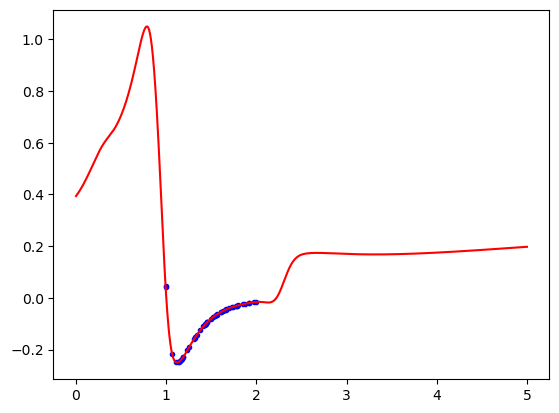
KANs do not save us from extrapolation issues though. I think a downside of KANs is it is not obvious what extrapolation behavior to expect. I guess it could be related to what happens in the spline representation of the functions. Eventually those have to extrapolate too.
X = torch.linspace(0, 5, 1000)[:, None] plt.plot(dataset['train_input'], dataset['train_label'], 'b.') plt.plot(X, model(X).detach().numpy(), 'r-');
It is early days for KANs, so many things we know about MLPs are still unknown for KANs. For example, with MLPs we know they extrapolate like the activation functions. Probably there is some insight like that to be had here, but it needs to be uncovered. With MLPs there are a lot of ways to regularize them for desired behavior. Probably that is true here too, and will be discovered. Similarly, there are many ways people have approached uncertainty quantification in MLPs that probably have some analog in KANs. Still, the ease of use suggests it could be promising for some applications.
Copyright (C) 2024 by John Kitchin. See the License for information about copying.
Org-mode version = 9.7-pre